From molecular diffusion dynamics to cellular function and beyond
I am a biochemist by training and excited to understand how molecular motion of bio-molecules translates to biological function across scales. It is utterly fascinating that the nano-scale interaction of only a few proteins such as transmembrane receptors binding their to ligands or transcription factors binding to DNA, can completely reshape the structure and function of a cell and even impact on whole tissues and organisms. To study these processes, I employ a variety of imaging, spectroscopy and molecular biology tools as well as in vitro model membrane systems, cell culture models, and animal models. Ultimately, I build tools and aim to answer fundamental questions about the dynamic organization of biological systems from the bottom up.
 Since March 2025, I am leading the Fluorescence and Membrane Dynamics (FMD) Lab at the University of Warwick. We are part of the Centre for Mechanochemical Cell Biology (CMCB) and the Cellular Interfaces Cluster. We are bringing together cell biologists, developmental biologists, and microscopists to understand the role of membrane organisation in the fundamental processes of life. Our main tools are fluorescence microscopy and spectroscopy and we are working hard to make every photon count.
Since March 2025, I am leading the Fluorescence and Membrane Dynamics (FMD) Lab at the University of Warwick. We are part of the Centre for Mechanochemical Cell Biology (CMCB) and the Cellular Interfaces Cluster. We are bringing together cell biologists, developmental biologists, and microscopists to understand the role of membrane organisation in the fundamental processes of life. Our main tools are fluorescence microscopy and spectroscopy and we are working hard to make every photon count.
My Research
At the heart of my research are the development, advancement, and application of fluorescence microscopy and spectroscopy methods to quantitatively follow cellular and sub-cellular dynamics.
Advancing fluorescence fluctuation spectroscopy
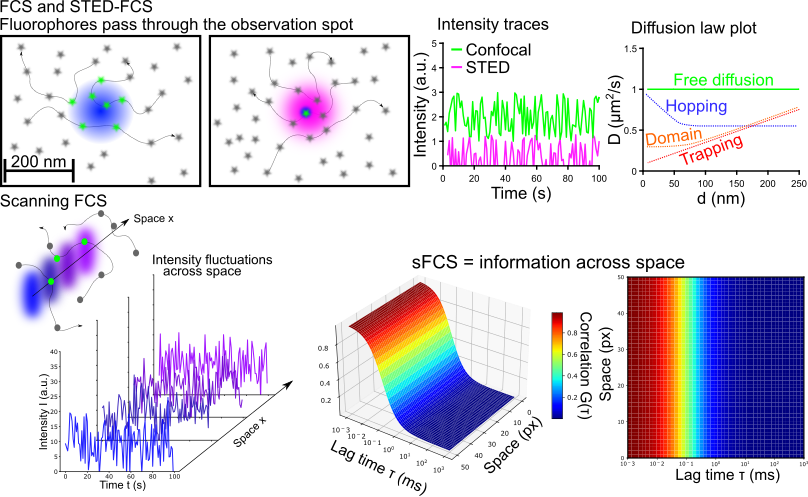
Fluorescence fluctuation spectroscopy (FFS) and especially fluorescence correlation spectroscopy (FCS) are commonly employed tools to study diffusion dynamics and changes in concentrations or oligomeric states. My research focuses on the application and further development of FCS in conjunction with super-resolution stimulated emission depletion (STED) microscopy. STED-FCS enables to measure diffusion dynamics directly on the nano-scale and even allows to map hindrances in diffusion (for example binding sites) across space. While STED-FCS provides unchallenged resolution for fluctuation spectroscopy, it requires special hardware. To make the nano-scale diffusion measurements more accessible, I work on confocal scanning FCS (sFCS) pipelines exploiting statistical tricks to access hidden dynamics in the measured ensemble. Currently, I am focussing my efforts to enable the detection of oligomerisation using sFCS.
Key Publications:
Schneider et al., Nature Comms (2024)
Schneider et al., J Phys D Appl Phys (2020)
Sezgin et al., Nat Protoc (2019)
Schneider et al., Nano Lett (2018)
Schneider et al., ACS Nano (2018)
Plasma membrane organisation & signalling
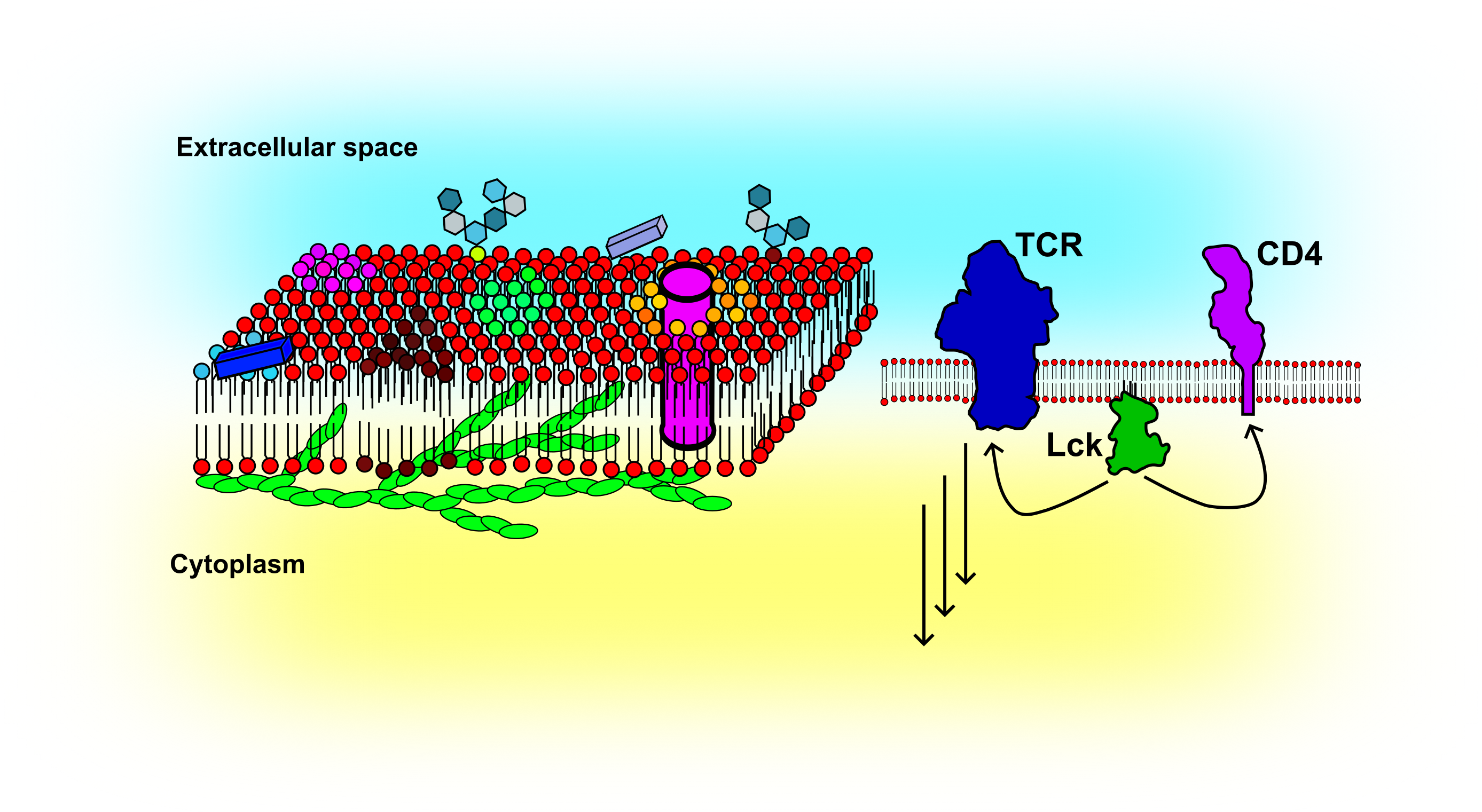
The cellular plasma membrane is composed of thousands of lipids and proteins. It acts as a signalling platform bringing molecules together or keeping them apart. The fluid nature of the membrane makes it a powerful cellular organizer. Dynamic techniques such as FCS can be applied to follow changes in, for example, lipid or receptor organization. Thus far, my research has focused on the impact of the actin cytoskeleton on the membrane organization and how FCS based methods can be used to reveal cellular activation in the context of immune cells. To study membrane organization, I routinely employ model membrane systems of different complexity such as supported lipid bilayers (SLBs), giant unilamellar vesicles (GUVs), giant plasma membrane vesicles (GPMVs), and cellular plasma membranes. More recently, I started to study membrane organization in the developing zebra fish embryo.
Key Publications:
Mørch et al., PNAS (2022)
Schneider et al., iScience (2021)
Pinkwart et al., J Biol Chem (2019)
Schneider et al., Mol Biol Cell (2017)
Biophysical imaging with smart probes
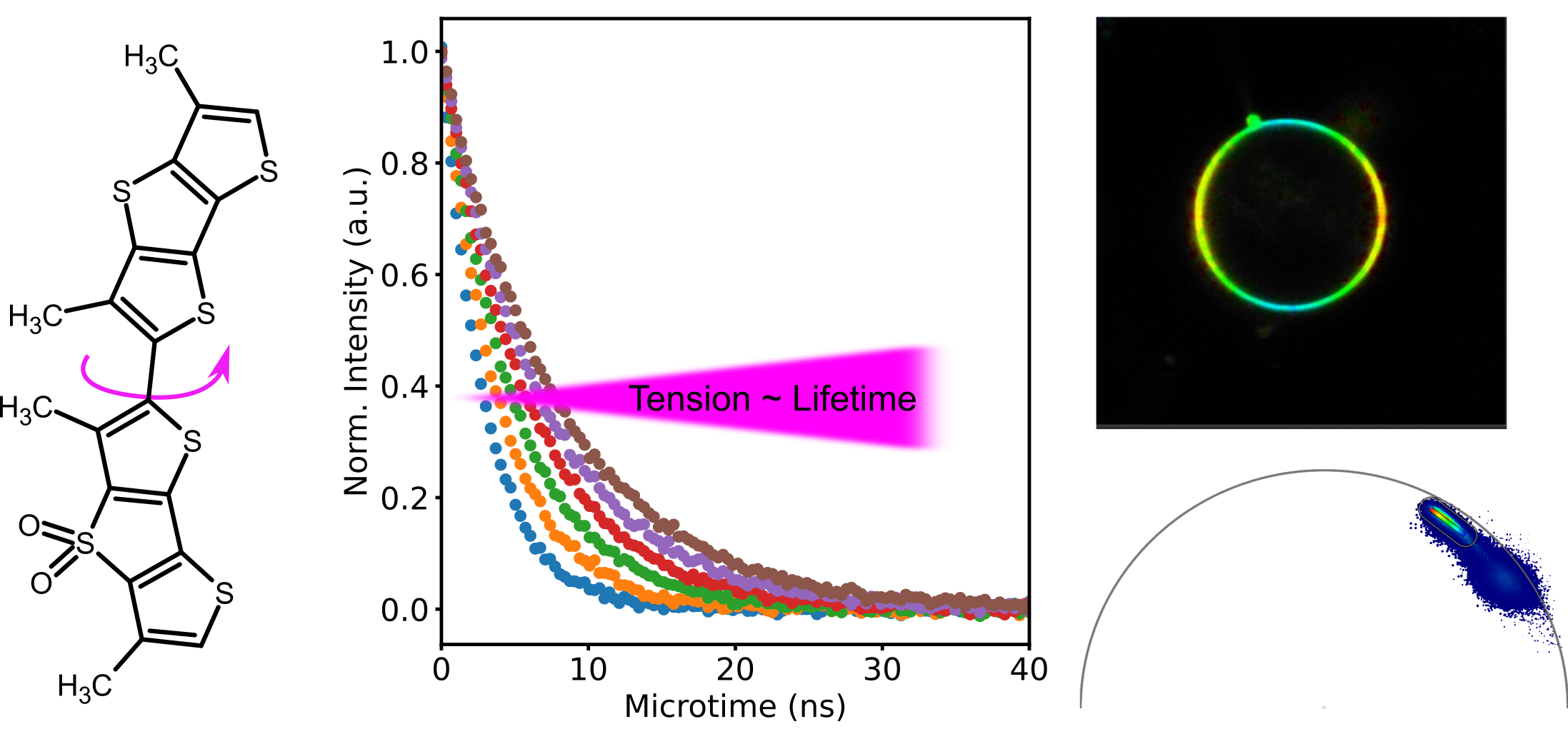
Cells are heterogeneous in their structure and organization: liquid condensates in the cytoplasm or nucleoplasm or nano-domain formation in the plasma membrane display only a small subset of possible means for the cell to compartmentalize signalling molecules. While measurements of diffusion dynamics can be used to infer organization, they are limited in spatial coverage. Smart probes, so called environment sensitive probes, overcome this issue. They can be used in imaging modality (with a large field of view) and change a certain fluorescence property according to the environment. For example, solvatochormic probes can report on lipid packing by a change in emission spectrum and can even be used in conjunction with super-resolution STED imaging. Another example is the Flipper probe that can report on membrane tension and is read out by changes in fluorescence lifetime. Recently, I have focussed on the use of the Flipper probe in in vivo settings such as mouse and zebra fish embryos and established that the dye can be efficiently used in conjunction with two-photon excitation. In addition, I have established the use of the Phasor analysis to aid visualisation of tension differneces.
Key Publications:
Roffay et al., Nat. Protoc. (2024)
Royer et al., Nat Commun (2022)
Sezgin et al., Biophys J (2017)
Simulations and data analysis
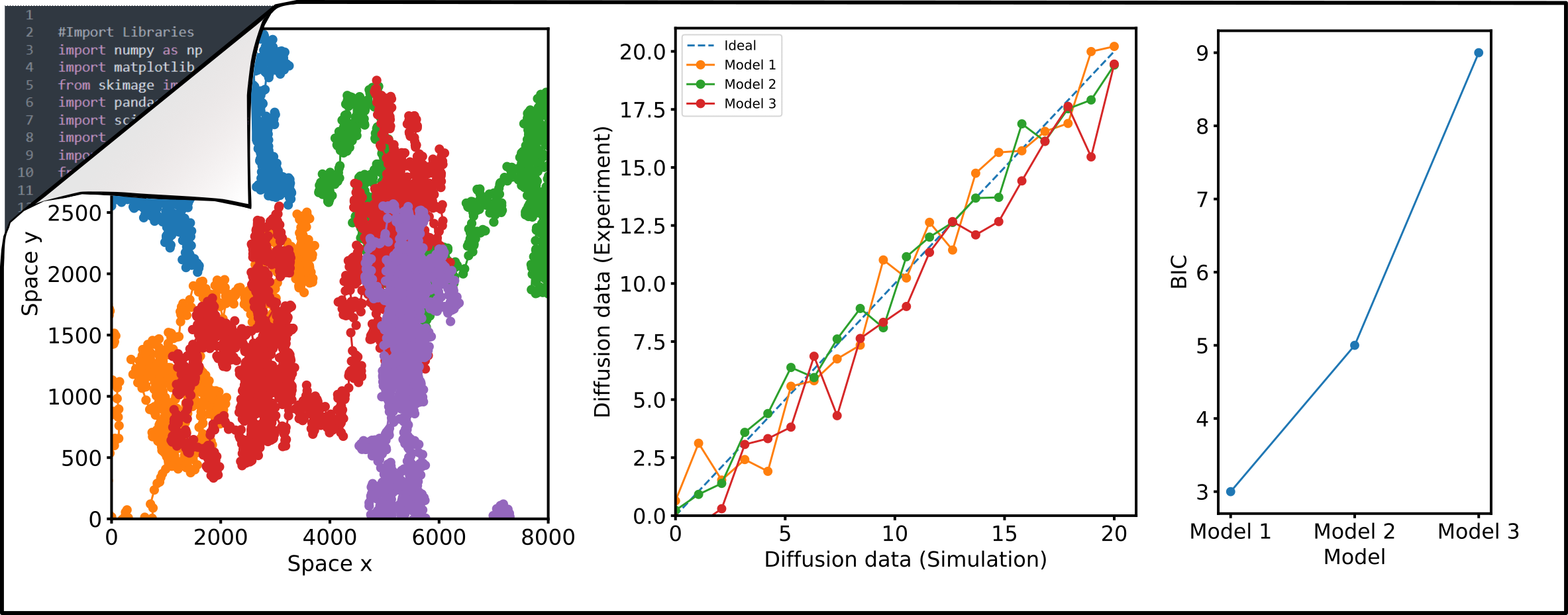
Numerical simulations of diffusion dynamics, diffusion modes, and oligomerization are invaluable as ground-truth for the evaluation of new approaches. I aim to develop robust analysis and simulation pipelines utilizing open-source Python programming that can be adapted and used by anyone. I have contributed to the development and continue to support the FoCuS_scan software package which provides a graphical user interface to analyse sFCS data. In addition, I write scripts for image analysis in FIJI for fast, automated, and most importantly reproducible image analysis. Codes are available through my Github page.
Key Publications:
Waithe et al., Methods (2018)
Schneider et al., ACS Nano (2018)
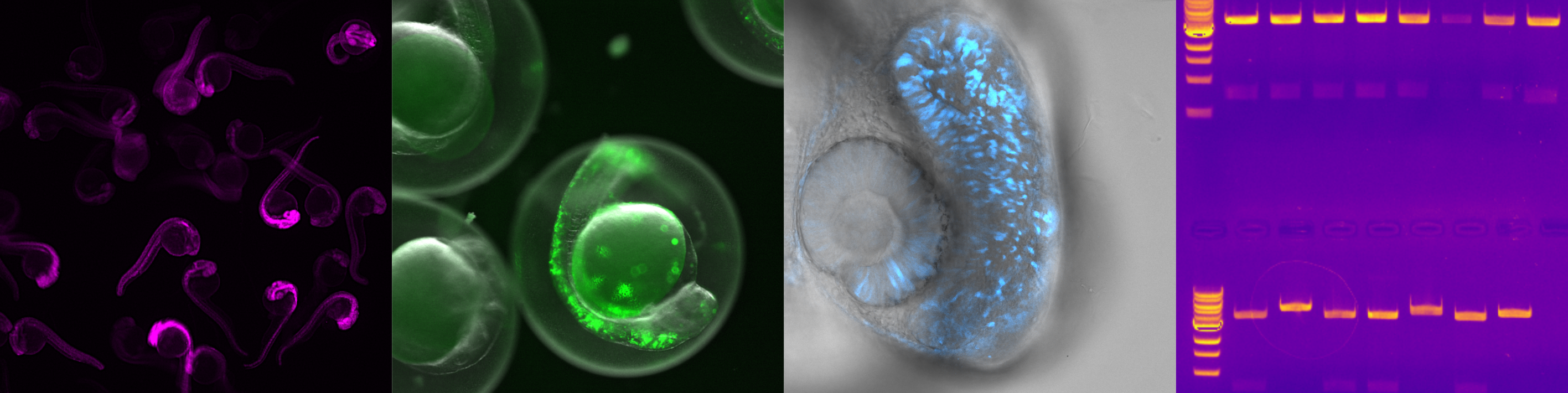
Studying molecular interactions in physiological context
Fundamental biological processes, including signal transduction, cellular differentiation, and cell migration, hinge upon single molecule interactions within living systems. Yet, these interactions are often studied in isolated conditions, such as using reconstituted proteins in solution or transfected cells in culture. To delve into more intricate biological questions, it is essential to investigate these interactions within the full physiological context, specifically within tissues and whole organisms. My recent research focuses on utilizing zebra fish as a model system to explore the influence of protein and nucleic acid interactions on early development. I employ standard and advanced molecular biology techniques for genetic modifications and have initiated the optimization of fluorescence fluctuation approaches and fluorescent probes for in vivo applications. This effort is geared towards enabling the observation of single molecule interactions using readily available instrumentation, facilitating the investigation of cellular signaling networks in developing tissues.
Key Publications:
Coming soon :)
CV
My academic CV can be downloaded here (coming soon)
My story in a few words
 Bachelor and Master Studies in Hanover
Bachelor and Master Studies in Hanover
In 2010, I started my scientific journey with the admission to the biochemistry program at the Leibniz University in Hanover, Germany. Choosing a field of study for the undergrad was quite challenging as I had a broad interest in natural sciences and math but also felt that the application and translation of basic science were important. The biochemistry program in Hanover was a collaboration between University and Medical school aiming to provide a broad basic science education combined with the prospect of medical applications, so it was a great fit for me. In addition, I had the chance to enjoy the life in a vibrant city offering a great mix of music scenes, history, and nature.
Throughout my studies and first lab experiences, I gained more and more interest in the phenomenon and application of fluorescence. The shear possibility of detecting a single molecule blew my mind and still amazes me. For my Bachelor thesis, I worked on the development of an inorganic phosphate sensor using microscale thermophoresis. During my master studies, I focused on biophysical chemistry and microscopy. I joined Christian Eggeling’s lab for a three-month internship at the University of Oxford which set the stage for my PhD. I was intrigued by the research and the upcoming field of super-resolution microscopy and spectroscopy. For my master thesis project, I joined Joerg Enderlein’s lab at the University of Göttingen which allowed me to learn more about spectroscopy and data analysis.
 PhD studies at Oxford
PhD studies at Oxford
I started my PhD at University of Oxford in the Eggeling Lab in October 2015. My project was based around the application of fluorescence fluctuation based methods in conjunction with super-resolution STED microscopy to shed light on the plasma membrane organization during T-cell activation. The Weatherall Institute for Molecular Medicine (WIMM) was the perfect place for the project because it brings together immunologists, cell biologists and microscopists. I thoroughly enjoyed the open and collaborative environment in the lab and building. I helped funding the Weatherall Graduate Student Association (WGSA) through which we organized frequently scientific and social events to further connect the student body.
Postdoctoral work at Oxford and USC
I defended my PhD in January 2020. While the Eggeling lab had moved to Jena, Germany, in 2018, I stayed for a transition postdoc in the Fritzsche Lab for Biophysical Immunology at the Kennedy Institute (University of Oxford). This allowed me to learn more about biomechanics and gave me time to conclude some projects from my PhD. Despite the COVID-19 pandemic, I could move to the US in May 2021 and started in Scott Fraser’s lab in the Translational Imaging Center (TIC) at the University of Southern California (USC). Currently, I am taking on the challenge of translating my quantitative imaging and spectroscopy tools to measurements in vivo, in zebrafish embryos, while also embracing life in Los Angeles.
Publications
All my published work can also be found through Google Scholar or ORCID.
Preprints - Manuscripts in Preparation
- Cherchia L, Fraser SE, Finley SD, Schneider F.
Prolactin receptor localization and dynamics: Insights from quantitative imaging and mathematical modeling
bioRxiv doi.org/10.1101/2025.10.14.682359. 2025
- Urbančič I, Schneider F, Galiani S, Sezgin E, Eggeling C.
Spectral STED microscopy improves spectral sensitivity with polarity-sensitive probes and enables correlative measurements of membrane order and anomalous lipid diffusion
bioRxiv doi.org/10.1101/2025.02.06.636942. 2025
- Stower M, Zhou F, Hathrell H, Yeung J, Thowfeequ S, Godwin J, Schneider F, Lagerholm BC, Fritzsche M, Thiyagalingam J, Lu X, Rittscher J, Srinivas S.
Single-cell phenomics reveals behavioural and mechanical heterogeneities underpinning collective migration during mouse anterior patterning
bioRxiv doi.org/10.1101/2023.03.31.534937. 2023
Published Research Articles
- Scott W, Ivorra-Molla E, Akhuli D, Massam-Wu T, Lysyganicz PK, Walsh R, Parent M, Cook J, Song L, Kumar A, Schneider F, Mishima M, Crow A, Balasubramanian M.
StayRose: a photostable StayGold derivative red-shifted by genetic code expansion
J Biol Chem. 2025
- Frei MS, Sanchez SA, He X, Liu L, Schneider F, Wang Z, Hakozaki H, Li Y, Lyons AC, Rohm TV, Olefsky JM, Shi L, Schoneberg J, Fraser SE, Mehta S, Wang Y, Zhang J.
Far-red chemigenetic kinase biosensors enable multiplexed and super-resolved imaging of signaling networks
Nat. Biotechnol.. 2025
-
Karedla N, Schneider F, Enderlein J, Chen T.
Leaflet-specific Structure and Dynamics of Solid and Polymer Supported Lipid Bilayers
Angew. Chem. Int. Ed., 1433-7851. 2025
-
Schneider F, Cespedes PF, Karedla N, Dustin ML, Fritzsche M.
Quantifying biomolecular organisation in membranes with brightness-transit statistics
Nat Commun 15, 7082. 2024
Featured by KIR/NDORMS News and RFI News
-
Mørch AM, Schneider F, Jenkins E, Santos AM, Fraser SE, Davis SJ, Dustin ML.
The kinase occupancy of T cell coreceptors reconsidered
PNAS 119 (49). 2022
Featured by phys.org
-
Galiani S, Reglinski K, Carravilla P, Barbotin A, Urbančič I, Ott J, Sehr J, Sezgin E, Schneider F, Waithe D, Hublitz P, Schliebs W, Erdmann R, Eggeling C.
Diffusion and interaction dynamics of the cytosolic peroxisomal import receptor PEX5
Biophys Rep 2(2). 2022
-
Royer C, Sandham E, Slee E, Schneider F, Lagerholm CB, Godwin J, Veits N, Hathrell H, Zhou F, Leonavicius K, Garratt J, Narendra T, Vincent A, Jones C, Child T, Coward K, Graham C, Fritzsche M, Lu X, Srinivas S.
ASPP2 maintains the integrity of mechanically stressed pseudostratified epithelia during morphogenesis
Nat Commun 13 941. 2022
-
Urbančič I, Schiffelers L, Jenkins E, Gong W, Santos AM, Schneider F, O'Brien-Ball C, Vuong MT, Ashman N, Sezgin E, Eggeling C.
Aggregation and mobility of membrane proteins interplay with local lipid order in the plasma membrane of T cells
FEBS Lett 595 16. 2021
-
Barbieri L, Colin-York H, Korobchevskaya K, Li D, Wolfson DL, Karedla N, Schneider F, Ahluwalia BS, Seternes T, Dalmo RA, Dustin ML, Li D, Fritzsche M.
Two-dimensional TIRF-SIM-traction force microscopy (2D TIRF-SIM-TFM)
Nat Commun 12 2169. 2021
-
Schneider F, Sych T, Eggeling C, Sezgin E.
Influence of nanobody binding on fluorescence emission, mobility, and organization of GFP-tagged proteins
iScience 24 1. 2021
-
Stanly TA, Fritzsche M, Banerji S, Shrestha D, Schneider F, Eggeling C, Jackson DG.
The cortical actin network regulates avidity-dependent binding of hyaluronan by the lymphatic vessel endothelial receptor LYVE-1
J Biol Chem 295 15. 2020
-
Schneider F, Hernandez-Varas P, Christoffer Lagerholm B, Shrestha D, Sezgin E, Julia Roberti M, Ossato G, Hecht F, Eggeling C, Urbančič I.
High photon count rates improve the quality of super-resolution fluorescence fluctuation spectroscopy
J Phys D Appl Phys 53 164003. 2020
-
Bedard M, Shrestha D, Priestman DA, Wang Y, Schneider F, Matute JD, Iyer SS, Gileadi U, Prota G, Kandasamy M, Veerapen N, Besra G, Fritzsche M, Zeissig S, Shevchenko A, Christianson JC, Platt FM, Eggeling C, Blumberg RS, Salio M, Cerundolo V.
Sterile activation of invariant natural killer T cells by ER-stressed antigen-presenting cells
PNAS 116 (47) 23671-23681. 2019
-
Pinkwart K, Schneider F, Lukoseviciute M, Sauka-Spengler T, Lyman E, Eggeling C, Sezgin E.
Nanoscale dynamics of cholesterol in the cell membrane
J Biol Chem 294 34. 2019
-
Sezgin E, Schneider F, Galiani S, Urbančič I, Waithe D, Lagerholm BC, Eggeling C.
Measuring nanoscale diffusion dynamics in cellular membranes with super-resolution STED-FCS
Nat Protoc 14 1054–1083. 2019
-
Urbančič I, Garvas M, Kokot B, Majaron H, Umek P, Cassidy H, Škarabot M, Schneider F, Galiani S, Arsov Z, Koklic T, Matallanas D, Čeh M, Muševič I, Eggeling C, Štrancar J.
Nanoparticles Can Wrap Epithelial Cell Membranes and Relocate Them Across the Epithelial Cell Layer
Nano Lett 18 8. 2018
-
Schneider F, Waithe D, Lagerholm BC, Shrestha D, Sezgin E, Eggeling C, Fritzsche M.
Statistical Analysis of Scanning Fluorescence Correlation Spectroscopy Data Differentiates Free from Hindered Diffusion
ACS Nano 12 8. 2018
-
Schneider F, Waithe D, Galiani S, Bernardino de la Serna J, Sezgin E, Eggeling C.
Nanoscale Spatiotemporal Diffusion Modes Measured by Simultaneous Confocal and Stimulated Emission Depletion Nanoscopy Imaging
Nano Lett 18 7. 2018
-
Waithe D, Schneider F, Chojnacki J, Clausen MP, Shrestha D, de la Serna JB, Eggeling C.
Optimized processing and analysis of conventional confocal microscopy generated scanning FCS data
Methods 140-141 62. 2018
-
Santos AM, Ponjavic A, Fritzsche M, Fernandes RA, de la Serna JB, Wilcock MJ, Schneider F, Urbančič I, McColl J, Anzilotti C, Ganzinger KA, Aßmann M, Depoil D, Cornall RJ, Dustin ML, Klenerman D, Davis SJ, Eggeling C, Lee SF.
Capturing resting T cells: the perils of PLL
Nat Immunol 19 203-205. 2018
-
Schneider F, Waithe D, Clausen MP, Galiani S, Koller T, Ozhan G, Eggeling C, Sezgin E.
Diffusion of lipids and GPI-anchored proteins in actin-free plasma membrane vesicles measured by STED-FCS
Mol Biol Cell 28 11. 2017
-
Sezgin E, Schneider F, Zilles V, Urbančič I, Garcia E, Waithe D, Klymchenko AS, Eggeling C.
Polarity-Sensitive Probes for Superresolution Stimulated Emission Depletion Microscopy
Biophys J. 113 6. 2017
-
Schneider F, Ruhlandt D, Gregor I, Enderlein J, Chizhik AI.
Quantum Yield Measurements of Fluorophores in Lipid Bilayers Using a Plasmonic Nanocavity
J Phys Chem Lett 8 7 1472. 2017
-
Clausen MP, Colin-York H, Schneider F, Eggeling C, Fritzsche M.
Dissecting the actin cortex density and membrane-cortex distance in living cells by super-resolution microscopy
J Phys D Appl Phys 50 064002. 2017
-
Steshenko O, Andrade DM, Honigmann A, Mueller V, Schneider F, Sezgin E, Hell SW, Simons M, Eggeling C.
Reorganization of Lipid Diffusion by Myelin Basic Protein as Revealed by STED Nanoscopy
Biophys J 110 11. 2016
-
Sezgin E, Can FB, Schneider F, Clausen MP, Galiani S, Stanly TA, Waithe D, Colaco A, Honigmann A, Wüstner D, Platt F, Eggeling C.
A comparative study on fluorescent cholesterol analogs as versatile cellular reporters
J Lipid Res 57 2 . 2016
Reviews and Book Chapters
-
Roffay C, García-Arcos JM, Chapuis P, López-Andarias J, Schneider F, Colom A, Tomba C, Di Meglio I, Barrett K, Dunsing V, Matile S, Roux A, Mercier V.
Tutorial: fluorescence lifetime microscopy of membrane mechanosensitive Flipper probes
Nat. Protoc. 2024
-
Luu P, Fraser SE, Schneider F.
More than double the fun with two-photon excitation microscopy
Commun. Biol. 7, 364. 2024
-
Mørch AM, Schneider F.
Investigating Diffusion Dynamics and Interactions with Scanning Fluorescence Correlation Spectroscopy (sFCS)
Chapter 5 in The Immune Synapse. Methods in Molecular Biology. 2023
-
Schneider F, Sezgin E.
Diffusion Measurements at the Nanoscale with STED-FCS
Fluorescence Spectroscopy and Microscopy in Biology. Springer Series on Fluorescence. 2022
-
Urbančič I, Lagerholm BC, Schneider F.
Fluorescence correlation spectroscopy
Chapter 33 in Imaging Modalities for Biological and Preclinical Research: A Compendium, Volume 1. 2021
-
Schneider F, Colin-York H, Fritzsche M.
Quantitative Bio-Imaging Tools to Dissect the Interplay of Membrane and Cytoskeletal Actin Dynamics in Immune Cells
Front Immunol 11 2020. 2021








